Accuracy of the AURORA test
The AURORA non-invasive prenatal test has shown a 99% specificity for all main fetal aneuploidies, with over 99.9% reliability in detecting the most common aneuploidies, i.e. Trisomy 21 and Trisomy 18. For Trisomy 13 and Monosomy X too, sensitivity rates are high (87.5% and 95%, respectively). In addition, for the most frequent trisomies the rate of false negatives and false positives is less than 0.1% and 0.3%, respectively. This indicates that, although the possibility of test error is extremely low, this, however, should not be entirely ruled out.
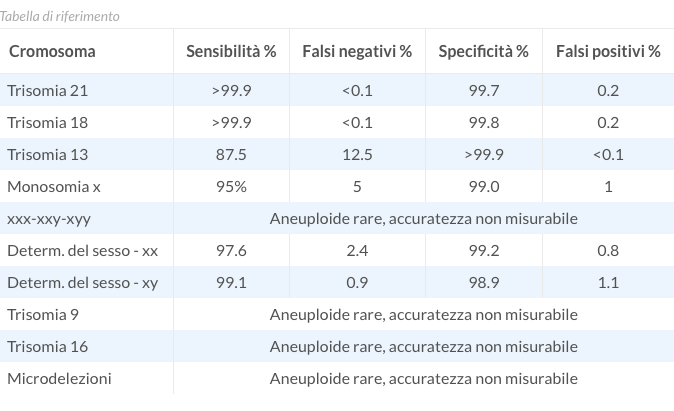
| Chromosome | Sensitivity % | False negatives % | Specificity % | False positives |
|---|---|---|---|---|
| Trisomy 21 | 99.14 | 0.86 | 99.94 | <0.1 |
| Trisomy 18 | 98.31 | 1.69 | 99.90 | 0.1 |
| Trisomy 13 | 98.15 | 1.85 | 99.95 | <0.1 |
| Monosomy x | 95 | 5 | 99.0 | 1 |
| xxx-xxy-xyy | Rare aneuploidies, unmeasurable accuracy | |||
| Sex determination - xx | 97.6 | 2.4 | 99.2 | 0.8 |
| Sex determination - xy | 99.1 | 0.9 | 98.9 | 1.1 |
| Trisomy 9 | Rare aneuploidies, unmeasurable accuracy | |||
| Trisomy 16 | Rare aneuploidies, unmeasurable accuracy | |||
| Microdeletions | Rare aneuploidies, unmeasurable accuracy | |||
Legend:
Sensitivity = power of the test, expressed in percentage terms, to identify the presence of a aneuploidy
Specificity = power of the test, expressed in percentage terms, to exclude those who have not developed an aneuploidy
False negatives = cases in which the test does not detect a chromosomal abnormality present in the fetus
False positives = cases in which the test detect a chromosomal abnormality which is not actually present in the fetus
*Bianchi DW et al., N Eng J Med 2014 Feb 27; 370(9):799-808
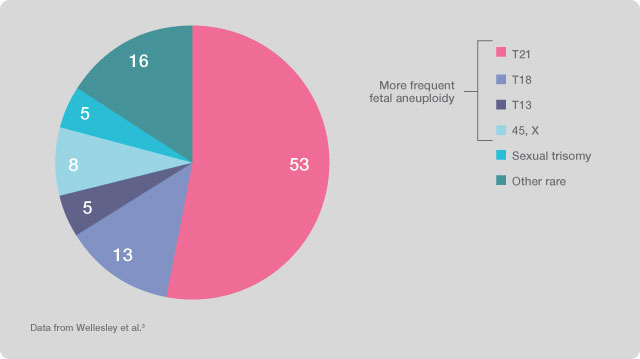
The validity of the results of this type of prenatal investigation has been verified and validated by a study on the test effectiveness, conducted by a team of researchers led by Dr. Diana W. Bianchi and published in Obstetrics and Gynaecology. The four main fetal chromosomal abnormalities identified by the tests account for about 80% of the total fetal chromosomal aneuploidies.
Figure 1. Prenatal incidence of chromosomal abnormalities
The massive parallel sequencing (MPS)of circulating cell free DNA (cfDNA) extracted from maternal plasma has proven to be an accurate and reliable method for chromosomal aneuploidy detection. The Normalized Chromosome Value (NCV) - the method used at Illumina labs - is calculated for each chromosome tested.
To support the validity of the test, there are several studies:
- Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901.
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105(42):16266–16271.
- Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem. 2011;57(7):1042–1049.
Since the publication of the first results of the works that validated the test effectiveness, Illumina researchers have been analyzing other cases, thus extending the case study and improving the test procedures and performance. These changes include a new DNA sequencing² chemistry, the optimization of the NCV calculation, the possibility of including the analysis of sexual chromosomes with six possible classifications and a redefinition of terminology to analyze chromosomes 21, 18 and 13 with a view to providing more accurate results.
To support the validity of non-invasive prenatal tests, it is possible to cite several studies:
- Prenatal detection of fetal triploidy from cell-free DNA testing in maternal blood. Nicolaides KH, Syngelaki A, del Mar Gil M, Soledad Quezada M, Zinevich Y. Fetal Diagn Ther 2013;doi:10.1159/000355655 *Externally-blinded, 20K protocol, NATUS, first study specifically aimed at triploidy detection.
- Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D. Prenat Diagn2013;33:1-5. *Externally-blinded, first published account of cfDNA-based triploidy detection, 20K protocol, NATUS.
- SNP-based non-invasive prenatal testing detects sex chromosome aneuploidies with high accuracy. Samango-Sprouse C, Banjevic M, Ryan A, et al. Prenat Diagn 2013; 33;643-9. *Sex chromosome aneuploidy detection, 20K protocol, NATUS.
- Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Zimmermann B, Hill M, Gemelos G, et al. Prenat Diagn 2012; 32:1233-41. *Proof-of-concept, 11K protocol, Parental Support.
- Non-invasive prenatal testing for whole chromosome abnormalities. Demko ZP, Zimmermann B, Rabinowitz M. J. Lab. Med. 2012;36:263-267.*Review.
- Cell-free fetal DNA and maternal serum analytes for monitoring embryonic and fetal status. Simpson JL. Fertil Steril 2013;99:1124–34. *Review.
- Non-invasive prenatal aneuploidy testing: technologies and clinical implications. Levy BL, Norwitz E. Med Lab Obs 2013. *Review.
- The Rapid Evolution of Noninvasive Prenatal Testing. Hall MP. IVD Technology 2013. *Review.
¹Bianchi et al., DNA Sequencing versus Standard Prenatal Aneuploidy Screening, NEJM 27.02.2014.
² Fan HC, Quake SR. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLoS One. 2010;5(5):e10439.